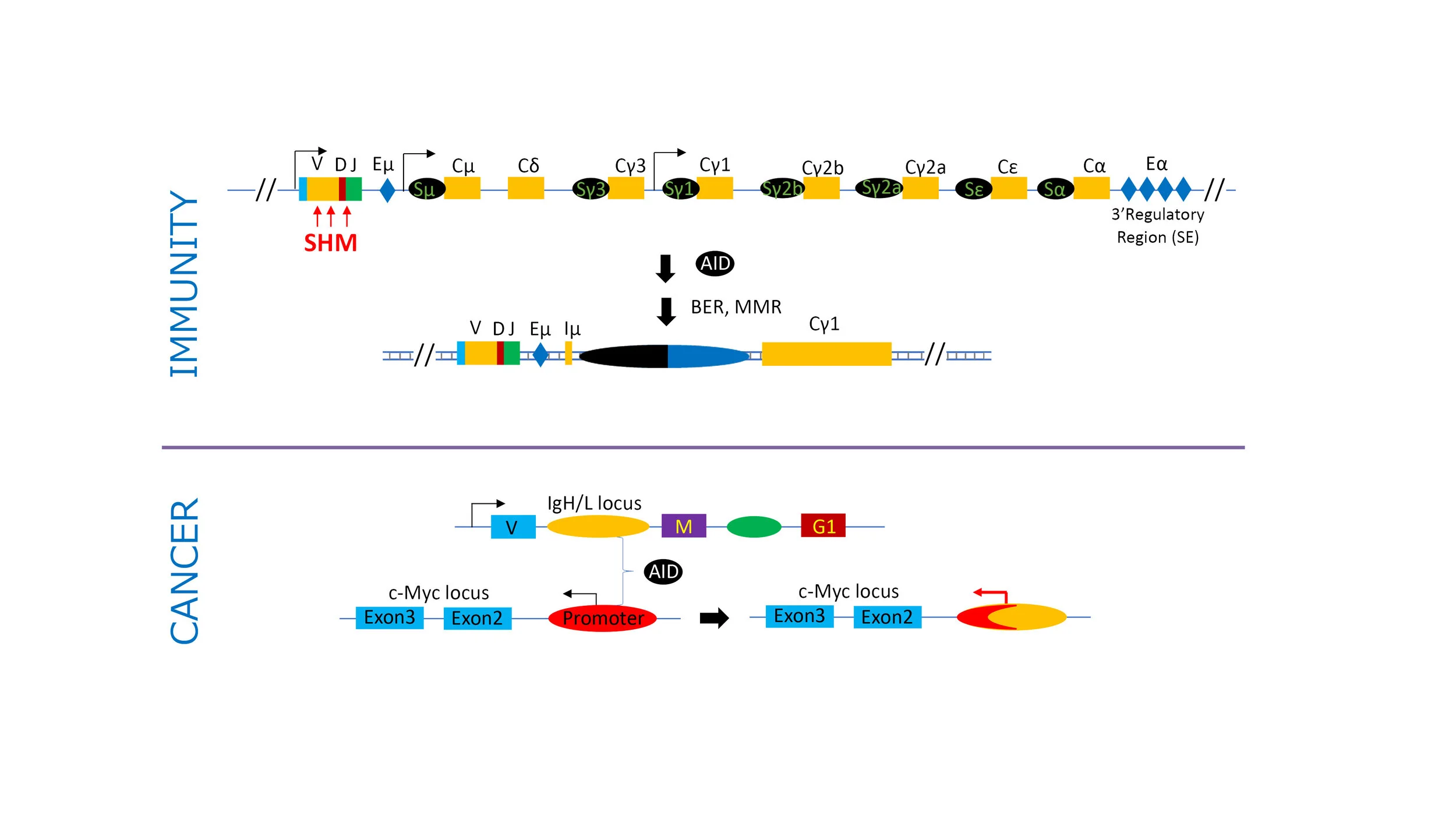
Somatic Hypermutation
Activation Induced cytidine Deaminase (AID), a member of the APOBEC family of DNA/RNA mutator enzymes, enhances the antibody repertoire by editing immunoglobulin locus sequences, driving class switch recombination (CSR) and somatic hypermutation (SHM). AID mediated mutations outside the Ig genes cause aberrant SHM and are associated with the cause or progression of B cell neoplasms. How SHM is restricted to the confinements of the Ig genes is defined by many parameters such as transcription regulation, epigenetics, topological organization of the genome, and DNA repair mechanisms. A mechanism integrating these factors is, however, lacking. The laboratory will continue to define the mechanisms that promote SHM to the Ig loci genes but suppress occurrence of oncogenic mutations in the rest of the B cell genome. Understanding physiological and aberrant SHM parameters will reward us in two ways: (1) unravel mechanism of affinity maturation of B cells through SHM for immune system’s ability to fight invading pathogens and (2) elucidate cell intrinsic mechanisms that cause and prevent mutations that have immediate implication in disease and far reaching consequence in evolution.
In additional to AID mediated Immunoglobulin locus mutagenesis events, our laboratory studies other aspects of genome mutagenesis that are caused at transcriptional regulatory elements like enhancers/super-enhancers and in context of topological organization of the genome.
RNA processing
Although vast regions of the mammalian genome are transcriptionally active, only a few RNAs are thought to be functionally relevant. We want to understand the life time of RNA as it encounters post/co-transcriptional modifications and undergoes surveillance via processing (3’ end decay) and degradative pathways. We generate animal models to tissue specifically ablate RNA modification and surveillance factors to understand various pathophysiological conditions that are seen in humans. We are expanding these studies to understand how the products of RNA processing can alter nuclear 3-dimensional chromosomal architecture as well as evaluating whether the products of processing have hitherto unexplored roles in the cell.
Our studies regarding noncoding RNA function and RNA processing focuses on biology of the immune system, neural system and reproductive system.